Ploutos | ||||
|
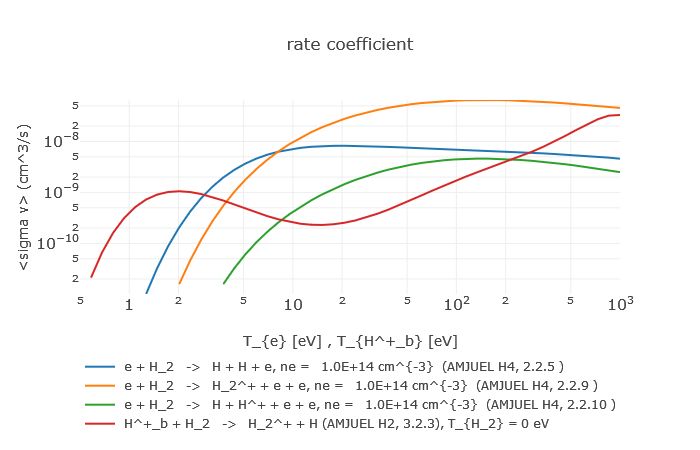
PLOUTOS is a data visualization and manipulation toolkit for EIRENE-related databases AMJUEL, HYDHEL and H2VIBR (Documentation). It has been developed from HYDKIN [Reiter09], a toolkit originally constructed for the data of hydro-carbon species, to allow easy data validation for the collisional-radiative models (CRMs) into EIRENE. PLOUTOS GUI provides the list of reaction data (cross sections, rates, population coefficients) in a interactive tabular form with • the same data from different databases/sections appearing in consecutive rows to fasten comparison between different sources, • retractable feature columns containing some useful information (reference, formula, comments, limits of validity, ..) to help the user tracing data origin and assessing data quality. The user selects a list of reaction data to plot (Figure 1) with the beam energy (for cross sections only) or the temperature range and other parameters (such as beam density or target energy) freely adjustable for sensitivity analysis.
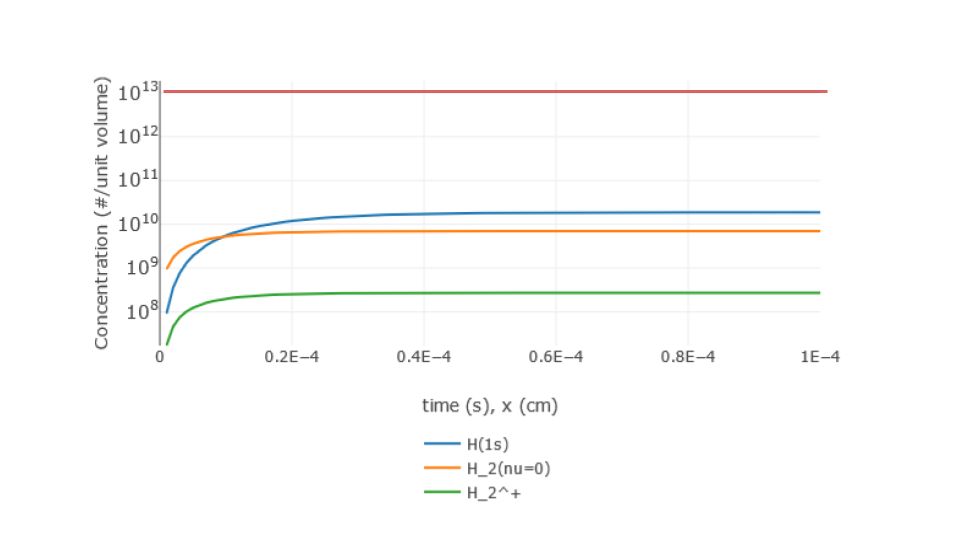
Alternatively, a CRM can be defined and the concentration of particle species is computed with a 1D solver, that allows to model a beam-like scenario and/or to perform linear spectral analysis (eigen-value approach) providing the relevant time-scale to equilibrium. The CRM is higly customizable by the user who can add particle sources, infinite reservoirs or finite 1D velocities, freeze some particles, introduce P and Q species, change background parameters (electron/ion/photon densities and temperatures). The outcome concentrations are plotted (Figure 2) for inspecting the results and can be re-used as initial conditions for next runs or downloaded in tabular/json formats for interaction with other codes.
PLOUTOS tools can be tested directly from the GUI on some default cases for molecular dissociation (with and without vibrational resolution) or by loading the json output of user’s previous runs. |